We’re in the midst of a paradigm shift in biomarker science. No longer academic luxuries or exploratory afterthoughts, biomarker signatures have evolved into strategic assets – tools that actively de-risk decision-making across the drug development continuum. From preclinical discovery to late-phase clinical trials, the right signature can streamline go/no-go calls, sharpen patient stratification, and validate mechanisms of action in real time.
This shift has been catalysed by two converging forces: the rise of high-dimensional, high-throughput platforms – from proteomics and transcriptomics to multiplexed imaging and single-cell technologies – and the integration of AI and advanced analytics into translational workflows. Together, they’ve opened the possibility of identifying signatures that are not only biologically rich, but also actionable, predictive, and modular.
Despite the growing availability of omics technologies and analytical platforms, many teams still struggle with a deceptively simple question: What kind of signature do we actually need?
Start with the clinical question, not the technology
Too often, the platform comes before the problem. A client might say, “We want to do proteomics on this study,” without asking: Why proteomics? What decision will this data inform?
Is the aim to identify patients most likely to respond? Do you need to prove target engagement? Are you trying to explain variability in response or disease trajectory? Each goal carries different requirements for timing, sample type, and resolution. And each approach comes with trade-offs – RNA is rich in pathway insight but less direct in function; proteins are closer to phenotype but can be limited by available detection reagents; cell-based assays give depth but are often logistically complex.
There’s also a tendency to want “one marker to rule them all.” But single-analyte biomarkers rarely perform in isolation. Biology is noisy, and redundancy is a feature, not a flaw. That’s why well-designed signatures are composite by design – combining multiple weak signals into a robust, interpretable readout.
So, how do you pick the right set of markers?
- Start with biological plausibility – include markers that make sense mechanistically.
- Ensure technical feasibility – are these markers reliably measurable in your chosen sample matrix, in a clinical setting, and at the volume and stability you can work with.
- Finally, ask whether they scale: can you validate these markers across cohorts, geographies, and analytical platforms?
The real art lies in choosing markers that are biologically relevant, clinically accessible, and analytically stable.
Platform convergence: When technologies de-risk each other
In translational science, there’s a seductive myth that one technology, one platform, or one dataset will tell us everything we need to know. But biology rarely cooperates with that level of simplicity. The most robust biomarker signatures aren’t built from a single source of truth; they emerge from convergence.
Platform convergence is the principle that different technologies, applied together, can resolve uncertainty, correct for each other’s blind spots, and strengthen confidence in a biological signal. Used strategically, technologies don’t just complement one another – they de-risk one another.
Let’s say a transcriptomic panel identifies a cluster of genes upregulated in responders to an immunotherapy. That’s interesting – but alone, it’s speculative. If those same patients also show:
- an increase in serum inflammatory proteins
- shifts in T cell subsets by flow cytometry, and
- activation markers visible on multiplexed tissue imaging,
– then you’re not looking at a hypothesis anymore. You’re looking at a mechanism in motion, triangulated by independent modalities. Each platform increases confidence in the others, and together, they reduce the risk of overinterpreting noise. This is how multi-omic signatures gain traction – not by being clever, but by being coherent.
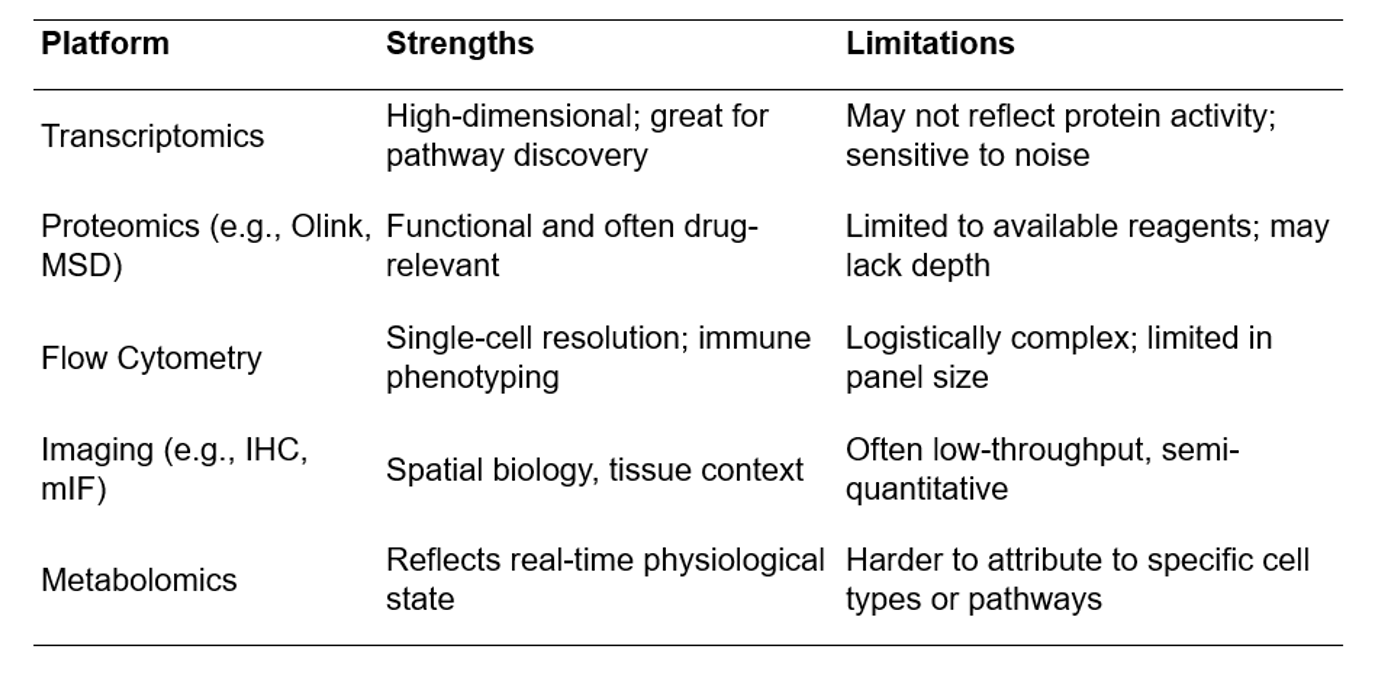
Strengths and Weaknesses: Why no single platform is enough
Platform design should mirror biological redundancy
Biology is redundant by nature. Cytokine signalling, for instance, often involves overlapping molecules and feedback loops. So why do we expect a single biomarker, or even a single technology, to capture that complexity? A resilient signature mimics this biological architecture: layered, flexible, and capable of generating a signal across variable conditions and sample quality. This doesn’t mean more noise, it means intentional overlap, where multiple markers or modalities speak to the same biological event from different angles. If IL-6 levels rise in a serum proteomics panel, you’d want to see correlating shifts in STAT3 pathway activation in transcriptomics, or expanded myeloid populations on flow cytometry. When these signals align, confidence increases. When they diverge, they prompt deeper investigation, sometimes uncovering new biology.
Redundant design also future-proofs a signature. Clinical trials are messy, samples get lost, sites vary, reagents drift. A signature that depends on a single fragile signal is brittle. But one built with overlapping layers can still yield insight even when part of the data is missing or noisy. This is especially critical in trials run across geographies, vendors, or long timelines.
Redundant design creates multiple entry points for assay simplification later. You may begin with a high-dimensional, multi-platform signature during early development, but through iterative refinement, reduce it to a handful of proteins or transcripts that still reflect the original biology – because you’ve mapped the redundancy and know what holds up.
Examples of Redundant Design in Action:
- Checkpoint blockade trials often rely on immune gene expression signatures (e.g., interferon-γ–driven profiles) that correlate with flow-based T cell activation and serum CXCL9/10 levels – reinforcing each signal.
- In fibrosis studies, protein-level markers of extracellular matrix remodelling (e.g., MMPs, TIMPs) are cross-validated with transcriptomic signatures from FFPE tissue and histological scoring – linking molecular change to tissue-level outcome.
- CAR-T programs may track both cytokine surges (Olink/MSD), immune cell dynamics (flow), and transcriptomic stress signatures (Nanostring or bulk RNA-seq) to build a robust safety and response-monitoring system.
Of course, not every study can afford all modalities up front. That’s where tiered platform strategies come in. A practical approach might look like this:
- Discovery: Use high-dimensional platforms (e.g., RNAseq, Olink Explore, scRNAseq) on a small subset.
- Refinement: Identify key features and reduce to targeted panels (e.g., flow, Olink Target 96).
- Validation: Apply refined, cost-effective assays across the broader trial population.
This approach doesn’t just save money – it preserves scientific integrity by ensuring that every phase of biomarker development is grounded in orthogonal evidence.
AI, complexity, and the clinically grounded signature
We’re now entering a phase of biomarker science where the limiting factor isn’t data – it’s interpretation. High-throughput platforms routinely produce datasets with tens of thousands of features, across dozens or hundreds of samples. AI and machine learning have become indispensable in sifting through this complexity to identify candidate signatures. But while these tools can accelerate discovery, they don’t replace biological insight. And they certainly don’t solve the challenge of clinical translation. AI can find patterns, but it can’t tell you what matters
The allure of AI lies in its ability to detect subtle, nonlinear patterns that would elude traditional statistical methods. Algorithms like random forests, LASSO regression, and support vector machines can identify multi-dimensional biomarker combinations that correlate with response, toxicity, or disease progression. But there’s a critical caveat: models are only as good as their input data, training logic, and interpretability. A model built on improperly normalized NPX values or inconsistently gated flow data will generate unreliable outputs – no matter how advanced the algorithm.
Even more crucial is clinical relevance. A signature composed of 50 transcriptomic features that require frozen biopsies and a GPU cluster to analyse may work brilliantly in a discovery dataset – but it’s dead on arrival in a real-world trial setting.
Interpretable, Actionable, and Portable
That’s why the next generation of biomarker signatures must meet a higher bar. They need to be:
- Interpretable: Clinicians and regulators must understand what the signature means.
- Actionable: It should drive a decision – stratify a patient, switch a dose, escalate care.
- Portable: The assay must be feasible in clinical trial conditions (e.g., in FFPE, serum, plasma; low sample volume; across geographies and lab vendors).
This is where the intersection of AI and domain expertise becomes powerful: human-guided feature selection combined with automated learning can yield simplified, robust signatures. A five-protein panel derived from an Olink study might ultimately outperform a 200-gene expression model if it’s more reproducible, easier to validate, and better aligned with clinical endpoints.
The operational litmus test: Is your signature deployable?
In our experience supporting biomarker development across diverse therapeutic areas and trial phases, we’ve seen how even the most biologically compelling signatures can stumble if operational feasibility isn’t considered from the outset. This isn’t a failure of science—it’s a misalignment of ambition and infrastructure.
The key is to design for the real-world constraints of clinical research. That means thinking beyond the discovery dataset to ensure your signature works across geographies, sample types, and clinical timelines.
Some guiding questions:
- Is the sample type (e.g., serum, CSF, FFPE) readily available in the study protocol?
- Can the assay be scaled to large, multi-site trials?
- Is there redundancy in the signature, in case of missing data?
- Can it be translated into a regulatory-grade assay later?
These are not minor details – they’re often the difference between exploratory data and a validated biomarker strategy.
Final Note: The signature is only as good as the system that supports it
An intelligent biomarker signature isn’t just an output – it’s part of a system. It must be traceable, reproducible, and capable of withstanding the rigors of GCP and regulatory scrutiny. That’s why the integration of AI and high-throughput data must be embedded into clinical trial operations, not running adjacent to them.
At Synexa, we believe the future of translational medicine belongs to companies that treat biomarker strategy not as a downstream deliverable, but as a core development discipline. We see biomarker signatures not as passive readouts, but as active design tools – shaping trials, clarifying decisions, and ultimately driving better outcomes for patients. We work closely with our partners to anticipate these critical factors early – ensuring that signatures are not only biologically meaningful, but also technically and logistically deployable. Whether supporting an early-phase mechanism-of-action study or a global Phase 3 trial, we emphasise that a successful signature is one that survives contact with the real world – and still delivers.
FAQ
What distinguishes a high‑value biomarker signature from a list of correlated features?
High‑value signatures are biologically coherent, reproducible across cohorts, and linked to actionable decisions such as dose, enrichment, or safety triage. Synexa prioritises mechanistic anchors, orthogonal confirmation, and assay deployability to prevent “overfit science.” Signatures are stress‑tested for platform, matrix, and operator variability before clinical embedding. This discipline ensures signatures survive real‑world trial conditions.
How do you operationalise signatures in multi‑centre studies without losing fidelity?
Method transfer packages, cross‑site training, and ring trials establish performance baselines, while unified data models keep outputs interoperable. Synexa’s QA systems enforce control materials, acceptance criteria, and drift monitoring with rapid remediation. Where feasible, central testing is used for complex modalities; otherwise, robust site‑readiness checks precede activation. This maintains signature integrity and interpretability.
How are signatures evolved over time without breaking comparability?
Versioning is governed: core markers remain stable while exploratory layers iterate under predefined rules. Synexa applies locked analysis pipelines with bridging datasets to compare “v1” and “v2” outputs directly. If improvements are material, controlled migration plans avoid loss of historical comparability. This lets programs benefit from innovation without resetting evidence.